Research Focus on Dr. F. Wayne Outten, Guy F. Lipscomb, Sr. Professor of Biochemistry, University of South Carolina
Dr. Nicholas Grossoehme, Professor of Chemistry, Winthrop University
Progress Towards Bio-inspired Drug Delivery Nanoparticles
Dr. Wayne Outten is a Guy F. Lipscomb, Sr. Professor of Biochemistry in the Department of Chemistry and Biochemistry at the University of South Carolina. Dr. Outten was recently elected as a 2020 Fellow of AAAS. Dr. Nicholas Grossoehme is a Professor of Chemistry in the Department of Chemistry, Physics, and Geology at Winthrop University. Their research contributes to the MADE in SC Thrust 2: Stimuli Responsive Polymeric Materials and Thrust 3: Rational Design of Interactive Biomaterials.
There are a number of different methods that are used in the delivery of drugs and therapeutics. Some of the more common methods include the oral consumption of pills and liquid medicines, intravenous injections, nasal and respiratory inhalation, and the topical or dermal application of ointments and creams. However, the problem with some of these methods is that they involve the systemic release of drugs throughout the entire body, typically resulting in lower potency at desired targets, in addition to unwanted side effects. As such, there is currently a large interest in the research and development of “smart” drug delivery systems (DDS) that can successfully circulate the blood and only release their contents at desired sites. This would result in (i) a controlled, localized release; (ii) higher drug concentrations and efficacy; and (iii) lower toxicity against non-targeted cells.
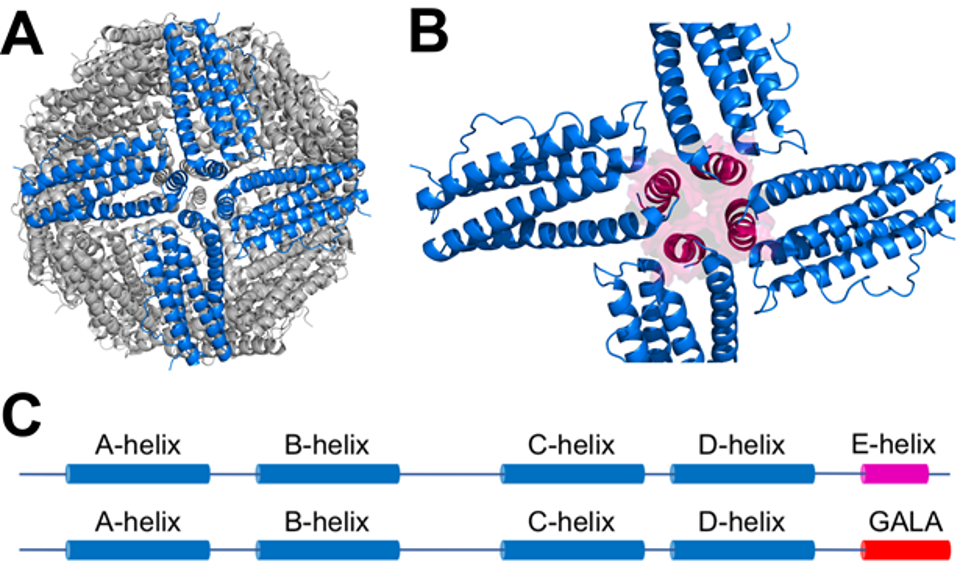
Figure 1. A. hFtnL with 4 monomer subunits along a 4-fold symmetry axis (with pore) highlighted in blue and other subunits in grey. B. Same 4 monomers magnified with the E-helix of each monomer shown in magenta as helix and transparent surface representation. Other monomers not shown. C. a-helical secondary structure (cylinders) in hFtnL (top) and hFtnL-GALA (bottom). Helix labels are per hFtnL nomenclature.
Ferritin, a ubiquitous protein used for intracellular iron storage and release, is an interesting candidate for use in drug delivery. Some of the individual characteristics that make it a reasonable contender include the following: a caged structure, thermal and pH stability, biocompatibility, the presence of a hollow cavity, dis- and reassembling capabilities, and genetic manipulability. For example, ferritin has been used to encapsulate anticancer drugs and has undergone genetic renovations to place targeting ligands on the outside of the ferritin shell to facilitate protein binding to the receptors of cancer cells and, subsequently, introducing the drug inside of the cell.
Ferritin is an oligomeric protein comprised of 24 subunits, which are a heterogenous combination of H (heavy) and/or L (light) chains. Each subunit consists of five α-helices (A-E) and an extended loop (BC loop) that connects the C-terminus of helix B with the N-terminus of helix C. Each subunit interacts such that helices A and C are oriented towards the outside of the protein shell, whereas helices B, D, and E are oriented towards the inside. Due to the elongation of helix D, the monomer chain folds back on itself and situates helix E at ~60° angle inside of the protein cage. This creates an integral 4-fold axis that is fundamental in the stability and functionality of ferritin. Figure 1B highlights the 4-fold axis and the positioning of the E-helix at the center of the axis.
Under neutral conditions, native ferritin spontaneously self-aggregates into an incredibly stable nanocage structure. Harshly acidic conditions (pH ~2.0 - 3.0) are required for the protein to disassemble, resulting in a variety of multimeric intermediates. Notably, hydrophobic interactions between amino acid residues of E-helices located at each of the 4-fold axes aid in the stabilization of this favorable conformation. It was previously shown that the native E-helix of hFtnL could be replaced with a peptide of repeating Glu-Ala-Glu-Ala (“GALA”) units, with His replacing Ala in the second repeat, without significantly altering the assembly of the Ftn 24-subunit nanocage at pH=7.0 (Figure 1C). At this pH the random coil secondary structure of GALA does not interfere with monomer subunit interactions around the 4-fold symmetry axis, although any subtle structural changes in that region have not been determined at the molecular level for hFtnL-GALA. Depending on the number of GALA repeats that are inserted, when the hFtnL-GALA nanocages are shifted to pH<6.0, a coil-helix transition in the GALA peptide causes disassembly of the nanocage into smaller subunits, presumably by disrupting monomer-monomer interactions at the 4-fold symmetry axis within the nanocage. The changes in hFtnL-GALA oligomeric state occur at pH regimes much higher than those required for native ferritin. The greater pH-responsiveness of hFtnL-GALA make it more useful as a biomaterial, especially for disassembly and delivery of cargo in the pH range 4.5 - 5.0 of the lysosome within the endosomal pathway. Although the hFtnL-GALA was partially characterized in vitro, the assembly/disassembly kinetics and thermodynamics have not been carefully analyzed, nor has it been structurally characterized beyond analyzing the oligomeric state in solution. There are no published attempts to load this pH-responsive nanocage with cargo, nor has the hFtnL-GALA been directly tested for interactions with native cell receptors (such as Scara5).
The goal of this project is the complete in vitro and in vivo characterization of the pH-responsive hFtnL-GALA nanocage in order to establish its usefulness as a stimuli-responsive biomaterial. The first step in this process was to purify the hFtnL-GALA peptide for further testing. Unexpectedly, the construct described in the literature did not express with high yields of soluble protein and the sample that was purified had limited solubility in the absence of imidazole and completely precipitated upon acidification below pH 7. These complications limited the progress toward the goals; however, the requirement for remote projects at Winthrop during most of the summer of 2020 offered an ideal opportunity for an in-silico undergraduate research project to critically evaluate the GALA-peptide, identify what properties were creating the pH-responsive transition and the solubility issues and attempt to design alternative peptides that retain the desirable features while addressing the solubility problem. Two main features were concluded from the evaluation of GALA.
- Under basic conditions, the random coil structure is favored due to electrostatic repulsion of the deprotonated Glu side chains. The coil-helix transition is nucleated by protonation of either His or Glu, which allows the formation of an H-bond with a Glu side chains three or four residues downstream. Consequently, the spacing of Glu residues is necessary to ensure pH responsiveness.
- The solubility issues originate from the high hydrophobic content of the GALA peptide. In helical form, there is likely an aggregation of helices into a coiled-coil structure which protects the hydrophobic side chains from water; however, at pH 7 where the nanocage is stable, the peptide adopts a random coil with no “protection” for the hydrophobic side chains. We hypothesized that replacing Leu with a more soluble amino acid would be ideal.
A variety of web-based applications were used to make predictions about the viability of GALA modifications: Jpred4 was used to predict the propensity to form an alpha helix and coiled-coil structure, H++ was used to predict side chain pKa values, and Peptide2.0 was used to assess the hydrophobicity and predict solubility. The threshold values used to identify viable candidates were > 90% propensity to form a coiled-coil and pKa values within 0.25 units of the GALA reference peptide. A broad variety of peptide combinations were screened. In general, we investigated how many Leu residues needed to be replaced, which hydrophilic amino acid or combination of amino acids, and the role of the Histidine residue present in the 2nd repeat. A number of attractive candidate peptides were identified with the most desirable traits originating from LeuàAsn or Gln substitutions.
To date, a series of six LeuàAsn proteins have been successfully cloned where one (EANA-1), two (EANA-2), three (EANA-3), four (EANA-4), five (EANA-5), or all six (EANA-6) Leu residues are replaced. EANA-6 has been purified and shows quite a bit of promise. The protein is readily accessible upon cell lysis and purifies in a simple process of metal affinity chromatography followed by size exclusion chromatography. The protein purifies to near homogeneity and can be concentrated to > 600 mM (> 13 mg/mL) in the absence of the imidazole stabilization that was required for the GALA variant. A pH-response is also observed; acidification below pH 6.8 triggers precipitation of the protein with only ~2% still soluble. Nearly 100% of the precipitated protein redissolves when the pH is adjusted back to 7.4 and analysis by size exclusion chromatography (Figure 2) strongly suggests that most of the protein reverts back to the nanocage structure. Notably, in the bottom panel of figure 2, a second peak appears at a retention volume of ~14.5 mL, indicating the presence of a smaller multimeric assembly of the EANA-6 protein. This observation is consistent with the acid-induced disassembly, generating insoluble multimeric intermediate(s). Adjusting the pH back to 7.4 induces the reassembly of the nanocage with a small percentage of intermediates unable to reassemble in the timeframe allowed.
In conclusion, our inability to reproduce the results expected based on the literature provided an excellent opportunity for an undergraduate research student to explore a different research path and see the value of in silico predictions to inform and focus in vitro experiments. Our recent research results also demonstrate the value of using computational, theory and modeling based approaches to inform experimental strategies in an iterative manner, which is a fundamental part of the “Materials Genome Initiative” of the MADE in SC EPSCoR.
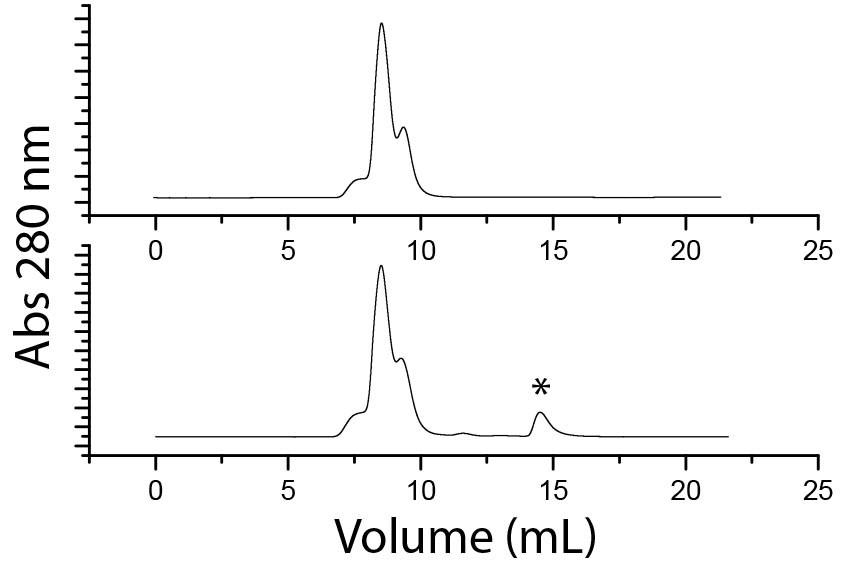
Figure 2 Superdex 10/300 G200 size exclusion chromatogram of hFtnL-EANA6-6 at pH 7.4 (top) and the resuspended precipitant following acid-induced precipitation of hFtnL-EANA6-6. The new peak (*) at a higher retention volume indicates a smaller multimeric aggregate of the protein that does not reassemble.
January 2021

